Build your own training/testing split
Date: 2021.10.07
When working with machine learning data, splitting into a “train”, “dev” (or validation) and “test”) set is important. Models use train data to learn representations and update their parameters; dev or validation data is reserved to see how the model may perform on unknown predictions. While it may not be explicitly trained on, it can be used as a stopping criteria, for hyper-parameter tuning, or as a simple sanity check. Lastly, test data is always reserved, hidden from the model, as a final pass to see what models perform best.
Lightwood supports a variety of encoders (Feature engineering procedures) and mixers (predictor algorithms that go from feature vectors to the target). Given the diversity of algorithms, it is appropriate to split data into these three categories when preparing encoders or fitting mixers.
Our default approach stratifies labeled data to ensure your train, validation, and test sets are equally represented in all classes. However, in many instances you may want a custom technique to build your own splits. We’ve included the splitter functionality (default found in lightwood.data.splitter) to enable you to build your own.
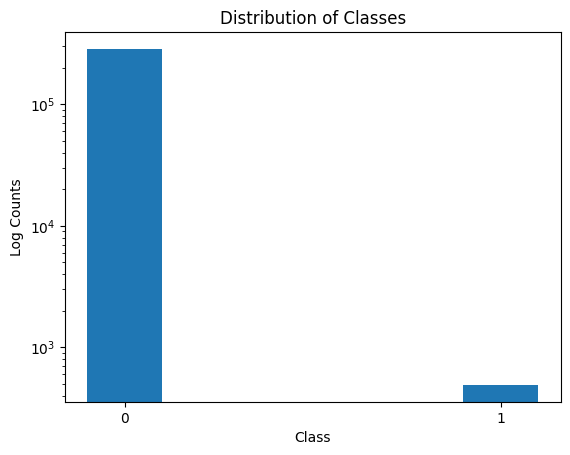
In the following problem, we shall work with a Kaggle dataset around credit card fraud (found here). Fraud detection is difficult because the events we are interested in catching are thankfully rare events. Because of that, there is a large imbalance of classes (in fact, in this dataset, less than 1% of the data are the rare-event).
In a supervised technique, we may want to ensure our training data sees the rare event of interest. A random shuffle could potentially miss rare events. We will implement SMOTE to increase the number of positive classes in our training data.
Let’s get started!
[1]:
import numpy as np
import pandas as pd
import torch
import nltk
import matplotlib.pyplot as plt
import os
import sys
# Lightwood modules
import lightwood as lw
from lightwood import ProblemDefinition, \
JsonAI, \
json_ai_from_problem, \
code_from_json_ai, \
predictor_from_code
import imblearn # Vers 0.5.0 minimum requirement
INFO:lightwood-3005:No torchvision detected, image helpers not supported.
INFO:lightwood-3005:No torchvision/pillow detected, image encoder not supported
1) Load your data
Lightwood works with pandas DataFrames. We can use pandas to load our data. Please download the dataset from the above link and place it in a folder called data/ where this notebook is located.
[2]:
# Load the data
data = pd.read_csv("https://mindsdb-example-data.s3.eu-west-2.amazonaws.com/jupyter/creditcard.csv.zip")
data.head()
[2]:
| Time | V1 | V2 | V3 | V4 | V5 | V6 | V7 | V8 | V9 | ... | V21 | V22 | V23 | V24 | V25 | V26 | V27 | V28 | Amount | Class | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.0 | -1.359807 | -0.072781 | 2.536347 | 1.378155 | -0.338321 | 0.462388 | 0.239599 | 0.098698 | 0.363787 | ... | -0.018307 | 0.277838 | -0.110474 | 0.066928 | 0.128539 | -0.189115 | 0.133558 | -0.021053 | 149.62 | 0 |
| 1 | 0.0 | 1.191857 | 0.266151 | 0.166480 | 0.448154 | 0.060018 | -0.082361 | -0.078803 | 0.085102 | -0.255425 | ... | -0.225775 | -0.638672 | 0.101288 | -0.339846 | 0.167170 | 0.125895 | -0.008983 | 0.014724 | 2.69 | 0 |
| 2 | 1.0 | -1.358354 | -1.340163 | 1.773209 | 0.379780 | -0.503198 | 1.800499 | 0.791461 | 0.247676 | -1.514654 | ... | 0.247998 | 0.771679 | 0.909412 | -0.689281 | -0.327642 | -0.139097 | -0.055353 | -0.059752 | 378.66 | 0 |
| 3 | 1.0 | -0.966272 | -0.185226 | 1.792993 | -0.863291 | -0.010309 | 1.247203 | 0.237609 | 0.377436 | -1.387024 | ... | -0.108300 | 0.005274 | -0.190321 | -1.175575 | 0.647376 | -0.221929 | 0.062723 | 0.061458 | 123.50 | 0 |
| 4 | 2.0 | -1.158233 | 0.877737 | 1.548718 | 0.403034 | -0.407193 | 0.095921 | 0.592941 | -0.270533 | 0.817739 | ... | -0.009431 | 0.798278 | -0.137458 | 0.141267 | -0.206010 | 0.502292 | 0.219422 | 0.215153 | 69.99 | 0 |
5 rows × 31 columns
We see 31 columns, most of these columns appear numerical. Due to confidentiality reasons, the Kaggle dataset mentions that the columns labeled \(V_i\) indicate principle components (PCs) from a PCA analysis of the original data from the credit card company. There is also a “Time” and “Amount”, two original features that remained. The time references time after the first transaction in the dataset, and amount is how much money was considered in the transaction.
You can also see a heavy imbalance in the two classes below:
[3]:
f = plt.figure()
ax = f.add_subplot(1,1,1)
ax.hist(data['Class'], bins = [-0.1, 0.1, 0.9, 1.1], log=True)
ax.set_ylabel("Log Counts")
ax.set_xticks([0, 1])
ax.set_xticklabels(["0", "1"])
ax.set_xlabel("Class")
ax.set_title("Distribution of Classes")
[3]:
Text(0.5, 1.0, 'Distribution of Classes')
2) Create a JSON-AI default object
We will now create JSON-AI syntax for our problem based on its specifications. We can do so by setting up a ProblemDefinition. The ProblemDefinition allows us to specify the target, the column we intend to predict, along with other details.
The end goal of JSON-AI is to provide **a set of instructions on how to compile a machine learning pipeline*.
Our target here is called “Class”, which indicates “0” for no fraud and “1” for fraud. We’ll generate the JSON-AI with the minimal syntax:
[4]:
# Setup the problem definition
problem_definition = {
'target': 'Class',
}
# Generate the j{ai}son syntax
json_ai = json_ai_from_problem(data, problem_definition)
INFO:type_infer-3005:Analyzing a sample of 18424
INFO:type_infer-3005:from a total population of 284807, this is equivalent to 6.5% of your data.
INFO:type_infer-3005:Using 3 processes to deduct types.
INFO:type_infer-3005:Infering type for: Time
INFO:type_infer-3005:Infering type for: V3
INFO:type_infer-3005:Infering type for: V6
INFO:type_infer-3005:Column Time has data type integer
INFO:type_infer-3005:Infering type for: V1
INFO:type_infer-3005:Column V3 has data type float
INFO:type_infer-3005:Infering type for: V4
INFO:type_infer-3005:Column V6 has data type float
INFO:type_infer-3005:Infering type for: V7
INFO:type_infer-3005:Column V4 has data type float
INFO:type_infer-3005:Infering type for: V5
INFO:type_infer-3005:Column V1 has data type float
INFO:type_infer-3005:Infering type for: V2
INFO:type_infer-3005:Column V5 has data type float
INFO:type_infer-3005:Infering type for: V9
INFO:type_infer-3005:Column V7 has data type float
INFO:type_infer-3005:Infering type for: V8
INFO:type_infer-3005:Column V9 has data type float
INFO:type_infer-3005:Infering type for: V10
INFO:type_infer-3005:Column V2 has data type float
INFO:type_infer-3005:Infering type for: V12
INFO:type_infer-3005:Column V10 has data type float
INFO:type_infer-3005:Infering type for: V11
INFO:type_infer-3005:Column V8 has data type float
INFO:type_infer-3005:Infering type for: V15
INFO:type_infer-3005:Column V12 has data type float
INFO:type_infer-3005:Infering type for: V13
INFO:type_infer-3005:Column V11 has data type float
INFO:type_infer-3005:Infering type for: V18
INFO:type_infer-3005:Column V15 has data type float
INFO:type_infer-3005:Infering type for: V16
INFO:type_infer-3005:Column V18 has data type float
INFO:type_infer-3005:Infering type for: V19
INFO:type_infer-3005:Column V13 has data type float
INFO:type_infer-3005:Infering type for: V14
INFO:type_infer-3005:Column V19 has data type float
INFO:type_infer-3005:Infering type for: V20
INFO:type_infer-3005:Column V16 has data type float
INFO:type_infer-3005:Infering type for: V17
INFO:type_infer-3005:Column V20 has data type float
INFO:type_infer-3005:Infering type for: V21
INFO:type_infer-3005:Column V14 has data type float
INFO:type_infer-3005:Infering type for: V24
INFO:type_infer-3005:Column V17 has data type float
INFO:type_infer-3005:Column V21 has data type float
INFO:type_infer-3005:Infering type for: V22
INFO:type_infer-3005:Infering type for: V27
INFO:type_infer-3005:Column V24 has data type float
INFO:type_infer-3005:Infering type for: V25
INFO:type_infer-3005:Column V22 has data type float
INFO:type_infer-3005:Infering type for: V23
INFO:type_infer-3005:Column V27 has data type float
INFO:type_infer-3005:Infering type for: V28
INFO:type_infer-3005:Column V23 has data type float
INFO:type_infer-3005:Infering type for: Class
INFO:type_infer-3005:Column Class has data type binary
INFO:type_infer-3005:Column V25 has data type float
INFO:type_infer-3005:Infering type for: V26
INFO:type_infer-3005:Column V28 has data type float
INFO:type_infer-3005:Infering type for: Amount
INFO:type_infer-3005:Column V26 has data type float
INFO:type_infer-3005:Column Amount has data type float
INFO:dataprep_ml-3005:Starting statistical analysis
INFO:dataprep_ml-3005:Finished statistical analysis
Lightwood looks at each of the many columns and indicates they are mostly float, with exception of “Class” which is binary.
You can observe the JSON-AI if you run the command print(json_ai.to_json()). Given there are many input features, we won’t print it out.
These are the only elements required to get off the ground with JSON-AI. However, we’re interested in making a custom approach. So, let’s make this syntax a file, and introduce our own changes.
3) Build your own splitter module
For Lightwood, the goal of a splitter is to intake an initial dataset (pre-processed ideally, although you can run the pre-processor on each DataFrame within the splitter) and return a dictionary with the keys “train”, “test”, and “dev” (at minimum). Subsequent steps of the pipeline expect the keys “train”, “test”, and “dev”, so it’s important you assign datasets to these as necessary.
We’re going to introduce SMOTE sampling in our splitter. SMOTE allows you to quickly learn an approximation to make extra “samples” that mimic the undersampled class.
We will use the package imblearn and scikit-learn to quickly create a train/test split and apply SMOTE to our training data only.
NOTE This is simply an example of things you can do with the splitter; whether SMOTE sampling is ideal for your problem depends on the question you’re trying to answer!
[5]:
%%writefile MyCustomSplitter.py
from type_infer.dtype import dtype
import pandas as pd
import numpy as np
from typing import List, Dict
from itertools import product
from lightwood.api.types import TimeseriesSettings
from lightwood.helpers.log import log
from imblearn.over_sampling import SMOTE
from sklearn.model_selection import train_test_split
def MySplitter(
data: pd.DataFrame,
target: str,
pct_train: float = 0.8,
pct_dev: float = 0.1,
seed: int = 1,
) -> Dict[str, pd.DataFrame]:
"""
Custom splitting function
:param data: Input data
:param target: Name of the target
:param pct_train: Percentage of data reserved for training, taken out of full data
:param pct_dev: Percentage of data reserved for dev, taken out of train data
:param seed: Random seed for reproducibility
:returns: A dictionary containing the keys train, test and dev with their respective data frames.
"""
# Shuffle the data
data = data.sample(frac=1, random_state=seed).reset_index(drop=True)
# Split into feature columns + target
X = data.iloc[:, data.columns != target] # .values
y = data[target] # .values
# Create a train/test split
X2, X_test, y2, y_test = train_test_split(
X, y, train_size=pct_train, random_state=seed, stratify=data[target]
)
X_train, X_dev, y_train, y_dev = train_test_split(
X2, y2, test_size=pct_dev, random_state=seed, stratify=y2
)
# Create a SMOTE model and bump up underbalanced class JUST for train data
SMOTE_model = SMOTE(random_state=seed)
Xtrain_mod, ytrain_mod = SMOTE_model.fit_resample(X_train, y_train.ravel())
Xtrain_mod[target] = ytrain_mod
X_test[target] = y_test
X_dev[target] = y_dev
return {"train": Xtrain_mod, "test": X_test, "dev": X_dev}
Writing MyCustomSplitter.py
Place your custom module in ~/lightwood_modules
We automatically search for custom scripts in your ~/lightwood_modules path. Place your file there. Later, you’ll see when we autogenerate code, that you can change your import location if you choose.
[6]:
from lightwood import load_custom_module
load_custom_module('MyCustomSplitter.py')
4) Introduce your custom splitter in JSON-AI
Now let’s introduce our custom splitter. JSON-AI keeps a lightweight syntax but fills in many default modules (like splitting, cleaning).
For the custom cleaner, we’ll work by editing the “splitter” key. We will change properties within it as follows:
“module” - place the name of the function. In our case it will be “MyCustomCleaner.cleaner”
“args” - any keyword argument specific to your cleaner’s internals.
This will look as follows:
"splitter": {
"module": "MyCustomSplitter.MySplitter",
"args": {
"data": "data",
"target": "$target",
"pct_train": 0.8,
"pct_dev": 0.1,
"seed": 1
}
},
5) Generate Python code representing your ML pipeline
Now we’re ready to load up our custom JSON-AI and generate the predictor code!
We can do this by first reading in our custom json-syntax, and then calling the function code_from_json_ai.
[7]:
json_ai.splitter = {
"module": "MyCustomSplitter.MySplitter",
"args": {
"data": "data",
"target": "$target",
"pct_train": 0.8,
"pct_dev": 0.1,
"seed": 1
}
}
#Generate python code that fills in your pipeline
code = code_from_json_ai(json_ai)
print(code)
# Save code to a file (Optional)
with open('custom_splitter_pipeline.py', 'w') as fp:
fp.write(code)
import lightwood
from lightwood import __version__ as lightwood_version
from lightwood.analysis import *
from lightwood.api import *
from lightwood.data import *
from lightwood.encoder import *
from lightwood.ensemble import *
from lightwood.helpers.device import *
from lightwood.helpers.general import *
from lightwood.helpers.ts import *
from lightwood.helpers.log import *
from lightwood.helpers.numeric import *
from lightwood.helpers.parallelism import *
from lightwood.helpers.seed import *
from lightwood.helpers.text import *
from lightwood.helpers.torch import *
from lightwood.mixer import *
from dataprep_ml.insights import statistical_analysis
from dataprep_ml.cleaners import cleaner
from dataprep_ml.splitters import splitter
from dataprep_ml.imputers import *
from mindsdb_evaluator import evaluate_accuracies
from mindsdb_evaluator.accuracy import __all__ as mdb_eval_accuracy_metrics
import pandas as pd
from typing import Dict, List, Union, Optional
import os
from types import ModuleType
import importlib.machinery
import sys
import time
for import_dir in [
os.path.join(
os.path.expanduser("~/lightwood_modules"), lightwood_version.replace(".", "_")
),
os.path.join("/etc/lightwood_modules", lightwood_version.replace(".", "_")),
]:
if os.path.exists(import_dir) and os.access(import_dir, os.R_OK):
for file_name in list(os.walk(import_dir))[0][2]:
if file_name[-3:] != ".py":
continue
mod_name = file_name[:-3]
loader = importlib.machinery.SourceFileLoader(
mod_name, os.path.join(import_dir, file_name)
)
module = ModuleType(loader.name)
loader.exec_module(module)
sys.modules[mod_name] = module
exec(f"import {mod_name}")
class Predictor(PredictorInterface):
target: str
mixers: List[BaseMixer]
encoders: Dict[str, BaseEncoder]
ensemble: BaseEnsemble
mode: str
def __init__(self):
seed(1)
self.target = "Class"
self.mode = "inactive"
self.problem_definition = ProblemDefinition.from_dict(
{
"target": "Class",
"pct_invalid": 2,
"unbias_target": True,
"seconds_per_mixer": 42768.0,
"seconds_per_encoder": None,
"expected_additional_time": 68.89478945732117,
"time_aim": 259200,
"target_weights": None,
"positive_domain": False,
"timeseries_settings": {
"is_timeseries": False,
"order_by": None,
"window": None,
"group_by": None,
"use_previous_target": True,
"horizon": None,
"historical_columns": None,
"target_type": "",
"allow_incomplete_history": True,
"eval_incomplete": False,
"interval_periods": [],
},
"anomaly_detection": False,
"use_default_analysis": True,
"embedding_only": False,
"dtype_dict": {},
"ignore_features": [],
"fit_on_all": True,
"strict_mode": True,
"seed_nr": 1,
}
)
self.accuracy_functions = ["balanced_accuracy_score"]
self.identifiers = {}
self.dtype_dict = {
"Time": "integer",
"V1": "float",
"V2": "float",
"V3": "float",
"V4": "float",
"V5": "float",
"V6": "float",
"V7": "float",
"V8": "float",
"V9": "float",
"V10": "float",
"V11": "float",
"V12": "float",
"V13": "float",
"V14": "float",
"V15": "float",
"V16": "float",
"V17": "float",
"V18": "float",
"V19": "float",
"V20": "float",
"V21": "float",
"V22": "float",
"V23": "float",
"V24": "float",
"V25": "float",
"V26": "float",
"V27": "float",
"V28": "float",
"Amount": "float",
"Class": "binary",
}
self.lightwood_version = "25.3.3.3"
self.pred_args = PredictionArguments()
# Any feature-column dependencies
self.dependencies = {
"Class": [],
"Time": [],
"V1": [],
"V2": [],
"V3": [],
"V4": [],
"V5": [],
"V6": [],
"V7": [],
"V8": [],
"V9": [],
"V10": [],
"V11": [],
"V12": [],
"V13": [],
"V14": [],
"V15": [],
"V16": [],
"V17": [],
"V18": [],
"V19": [],
"V20": [],
"V21": [],
"V22": [],
"V23": [],
"V24": [],
"V25": [],
"V26": [],
"V27": [],
"V28": [],
"Amount": [],
}
self.input_cols = [
"Time",
"V1",
"V2",
"V3",
"V4",
"V5",
"V6",
"V7",
"V8",
"V9",
"V10",
"V11",
"V12",
"V13",
"V14",
"V15",
"V16",
"V17",
"V18",
"V19",
"V20",
"V21",
"V22",
"V23",
"V24",
"V25",
"V26",
"V27",
"V28",
"Amount",
]
# Initial stats analysis
self.statistical_analysis = None
self.ts_analysis = None
self.runtime_log = dict()
self.global_insights = dict()
# Feature cache
self.feature_cache = dict()
@timed_predictor
def analyze_data(self, data: pd.DataFrame) -> None:
# Perform a statistical analysis on the unprocessed data
self.statistical_analysis = statistical_analysis(
data, self.dtype_dict, self.problem_definition.to_dict(), {}
)
# Instantiate post-training evaluation
self.analysis_blocks = [
ICP(fixed_significance=None, confidence_normalizer=False, deps=[]),
ConfStats(deps=["ICP"]),
AccStats(deps=["ICP"]),
PermutationFeatureImportance(deps=["AccStats"]),
]
@timed_predictor
def preprocess(self, data: pd.DataFrame) -> pd.DataFrame:
# Preprocess and clean data
log.info("Cleaning the data")
self.imputers = {}
data = cleaner(
data=data,
pct_invalid=self.problem_definition.pct_invalid,
identifiers=self.identifiers,
dtype_dict=self.dtype_dict,
target=self.target,
mode=self.mode,
imputers=self.imputers,
timeseries_settings=self.problem_definition.timeseries_settings.to_dict(),
anomaly_detection=self.problem_definition.anomaly_detection,
)
# Time-series blocks
return data
@timed_predictor
def split(self, data: pd.DataFrame) -> Dict[str, pd.DataFrame]:
# Split the data into training/testing splits
log.info("Splitting the data into train/test")
train_test_data = MyCustomSplitter.MySplitter(
data=data, pct_train=0.8, pct_dev=0.1, seed=1, target=self.target
)
return train_test_data
@timed_predictor
def prepare(self, data: Dict[str, pd.DataFrame]) -> None:
# Prepare encoders to featurize data
self.mode = "train"
if self.statistical_analysis is None:
raise Exception("Please run analyze_data first")
# Column to encoder mapping
self.encoders = {
"Class": BinaryEncoder(
is_target=True, target_weights=self.statistical_analysis.target_weights
),
"Time": NumericEncoder(),
"V1": NumericEncoder(),
"V2": NumericEncoder(),
"V3": NumericEncoder(),
"V4": NumericEncoder(),
"V5": NumericEncoder(),
"V6": NumericEncoder(),
"V7": NumericEncoder(),
"V8": NumericEncoder(),
"V9": NumericEncoder(),
"V10": NumericEncoder(),
"V11": NumericEncoder(),
"V12": NumericEncoder(),
"V13": NumericEncoder(),
"V14": NumericEncoder(),
"V15": NumericEncoder(),
"V16": NumericEncoder(),
"V17": NumericEncoder(),
"V18": NumericEncoder(),
"V19": NumericEncoder(),
"V20": NumericEncoder(),
"V21": NumericEncoder(),
"V22": NumericEncoder(),
"V23": NumericEncoder(),
"V24": NumericEncoder(),
"V25": NumericEncoder(),
"V26": NumericEncoder(),
"V27": NumericEncoder(),
"V28": NumericEncoder(),
"Amount": NumericEncoder(),
}
# Prepare the training + dev data
concatenated_train_dev = pd.concat([data["train"], data["dev"]])
prepped_encoders = {}
# Prepare input encoders
parallel_encoding = parallel_encoding_check(data["train"], self.encoders)
if parallel_encoding:
log.debug("Preparing in parallel...")
for col_name, encoder in self.encoders.items():
if col_name != self.target and not encoder.is_trainable_encoder:
prepped_encoders[col_name] = (
encoder,
concatenated_train_dev[col_name],
"prepare",
)
prepped_encoders = mut_method_call(prepped_encoders)
else:
log.debug("Preparing sequentially...")
for col_name, encoder in self.encoders.items():
if col_name != self.target and not encoder.is_trainable_encoder:
log.debug(f"Preparing encoder for {col_name}...")
encoder.prepare(concatenated_train_dev[col_name])
prepped_encoders[col_name] = encoder
# Store encoders
for col_name, encoder in prepped_encoders.items():
self.encoders[col_name] = encoder
# Prepare the target
if self.target not in prepped_encoders:
if self.encoders[self.target].is_trainable_encoder:
self.encoders[self.target].prepare(
data["train"][self.target], data["dev"][self.target]
)
else:
self.encoders[self.target].prepare(
pd.concat([data["train"], data["dev"]])[self.target]
)
# Prepare any non-target encoders that are learned
for col_name, encoder in self.encoders.items():
if col_name != self.target and encoder.is_trainable_encoder:
priming_data = pd.concat([data["train"], data["dev"]])
kwargs = {}
if self.dependencies[col_name]:
kwargs["dependency_data"] = {}
for col in self.dependencies[col_name]:
kwargs["dependency_data"][col] = {
"original_type": self.dtype_dict[col],
"data": priming_data[col],
}
# If an encoder representation requires the target, provide priming data
if hasattr(encoder, "uses_target"):
kwargs["encoded_target_values"] = self.encoders[self.target].encode(
priming_data[self.target]
)
encoder.prepare(
data["train"][col_name], data["dev"][col_name], **kwargs
)
@timed_predictor
def featurize(self, split_data: Dict[str, pd.DataFrame]):
# Featurize data into numerical representations for models
log.info("Featurizing the data")
tss = self.problem_definition.timeseries_settings
feature_data = dict()
for key, data in split_data.items():
if key != "stratified_on":
# compute and store two splits - full and filtered (useful for time series post-train analysis)
if key not in self.feature_cache:
featurized_split = EncodedDs(self.encoders, data, self.target)
filtered_subset = EncodedDs(
self.encoders, filter_ts(data, tss), self.target
)
for k, s in zip(
(key, f"{key}_filtered"), (featurized_split, filtered_subset)
):
self.feature_cache[k] = s
for k in (key, f"{key}_filtered"):
feature_data[k] = self.feature_cache[k]
return feature_data
@timed_predictor
def fit(self, enc_data: Dict[str, pd.DataFrame]) -> None:
# Fit predictors to estimate target
self.mode = "train"
# --------------- #
# Extract data
# --------------- #
# Extract the featurized data into train/dev/test
encoded_train_data = enc_data["train"]
encoded_dev_data = enc_data["dev"]
encoded_test_data = enc_data["test_filtered"]
log.info("Training the mixers")
# --------------- #
# Fit Models
# --------------- #
# Assign list of mixers
self.mixers = [
Neural(
fit_on_dev=True,
search_hyperparameters=True,
net="DefaultNet",
stop_after=self.problem_definition.seconds_per_mixer,
target=self.target,
dtype_dict=self.dtype_dict,
target_encoder=self.encoders[self.target],
),
XGBoostMixer(
fit_on_dev=True,
use_optuna=True,
stop_after=self.problem_definition.seconds_per_mixer,
target=self.target,
dtype_dict=self.dtype_dict,
input_cols=self.input_cols,
target_encoder=self.encoders[self.target],
),
Regression(
stop_after=self.problem_definition.seconds_per_mixer,
target=self.target,
dtype_dict=self.dtype_dict,
target_encoder=self.encoders[self.target],
),
RandomForest(
fit_on_dev=True,
stop_after=self.problem_definition.seconds_per_mixer,
target=self.target,
dtype_dict=self.dtype_dict,
target_encoder=self.encoders[self.target],
),
]
# Train mixers
trained_mixers = []
for mixer in self.mixers:
try:
if mixer.trains_once:
self.fit_mixer(
mixer,
ConcatedEncodedDs([encoded_train_data, encoded_dev_data]),
encoded_test_data,
)
else:
self.fit_mixer(mixer, encoded_train_data, encoded_dev_data)
trained_mixers.append(mixer)
except Exception as e:
log.warning(f"Exception: {e} when training mixer: {mixer}")
if True and mixer.stable:
raise e
# Update mixers to trained versions
if not trained_mixers:
raise Exception(
"No mixers could be trained! Please verify your problem definition or JsonAI model representation."
)
self.mixers = trained_mixers
# --------------- #
# Create Ensembles
# --------------- #
log.info("Ensembling the mixer")
# Create an ensemble of mixers to identify best performing model
# Dirty hack
self.ensemble = BestOf(
data=encoded_test_data,
fit=True,
ts_analysis=None,
target=self.target,
mixers=self.mixers,
args=self.pred_args,
accuracy_functions=self.accuracy_functions,
)
self.supports_proba = self.ensemble.supports_proba
@timed_predictor
def fit_mixer(self, mixer, encoded_train_data, encoded_dev_data) -> None:
mixer.fit(encoded_train_data, encoded_dev_data)
@timed_predictor
def analyze_ensemble(self, enc_data: Dict[str, pd.DataFrame]) -> None:
# Evaluate quality of fit for the ensemble of mixers
# --------------- #
# Extract data
# --------------- #
# Extract the featurized data into train/dev/test
encoded_train_data = enc_data["train"]
encoded_dev_data = enc_data["dev"]
encoded_test_data = enc_data["test"]
# --------------- #
# Analyze Ensembles
# --------------- #
log.info("Analyzing the ensemble of mixers")
self.model_analysis, self.runtime_analyzer = model_analyzer(
data=encoded_test_data,
train_data=encoded_train_data,
ts_analysis=None,
stats_info=self.statistical_analysis,
pdef=self.problem_definition,
accuracy_functions=self.accuracy_functions,
predictor=self.ensemble,
target=self.target,
dtype_dict=self.dtype_dict,
analysis_blocks=self.analysis_blocks,
)
@timed_predictor
def learn(self, data: pd.DataFrame) -> None:
if self.problem_definition.ignore_features:
log.info(f"Dropping features: {self.problem_definition.ignore_features}")
data = data.drop(
columns=self.problem_definition.ignore_features, errors="ignore"
)
self.mode = "train"
n_phases = 8 if self.problem_definition.fit_on_all else 7
# Perform stats analysis
log.info(f"[Learn phase 1/{n_phases}] - Statistical analysis")
self.analyze_data(data)
# Pre-process the data
log.info(f"[Learn phase 2/{n_phases}] - Data preprocessing")
data = self.preprocess(data)
# Create train/test (dev) split
log.info(f"[Learn phase 3/{n_phases}] - Data splitting")
train_dev_test = self.split(data)
# Prepare encoders
log.info(f"[Learn phase 4/{n_phases}] - Preparing encoders")
self.prepare(train_dev_test)
# Create feature vectors from data
log.info(f"[Learn phase 5/{n_phases}] - Feature generation")
enc_train_test = self.featurize(train_dev_test)
# Prepare mixers
log.info(f"[Learn phase 6/{n_phases}] - Mixer training")
if not self.problem_definition.embedding_only:
self.fit(enc_train_test)
else:
self.mixers = []
self.ensemble = Embedder(
self.target, mixers=list(), data=enc_train_test["train"]
)
self.supports_proba = self.ensemble.supports_proba
# Analyze the ensemble
log.info(f"[Learn phase 7/{n_phases}] - Ensemble analysis")
self.analyze_ensemble(enc_train_test)
# ------------------------ #
# Enable model partial fit AFTER it is trained and evaluated for performance with the appropriate train/dev/test splits.
# This assumes the predictor could continuously evolve, hence including reserved testing data may improve predictions.
# SET `json_ai.problem_definition.fit_on_all=False` TO TURN THIS BLOCK OFF.
# Update the mixers with partial fit
if self.problem_definition.fit_on_all and all(
[not m.trains_once for m in self.mixers]
):
log.info(f"[Learn phase 8/{n_phases}] - Adjustment on validation requested")
self.adjust(
enc_train_test["test"].data_frame,
ConcatedEncodedDs(
[enc_train_test["train"], enc_train_test["dev"]]
).data_frame,
adjust_args={"learn_call": True},
)
self.feature_cache = (
dict()
) # empty feature cache to avoid large predictor objects
@timed_predictor
def adjust(
self,
train_data: Union[EncodedDs, ConcatedEncodedDs, pd.DataFrame],
dev_data: Optional[Union[EncodedDs, ConcatedEncodedDs, pd.DataFrame]] = None,
adjust_args: Optional[dict] = None,
) -> None:
# Update mixers with new information
self.mode = "train"
# --------------- #
# Prepare data
# --------------- #
if dev_data is None:
data = train_data
split = splitter(
data=data,
pct_train=0.8,
pct_dev=0.2,
pct_test=0,
tss=self.problem_definition.timeseries_settings.to_dict(),
seed=self.problem_definition.seed_nr,
target=self.target,
dtype_dict=self.dtype_dict,
)
train_data = split["train"]
dev_data = split["dev"]
if adjust_args is None or not adjust_args.get("learn_call"):
train_data = self.preprocess(train_data)
dev_data = self.preprocess(dev_data)
dev_data = EncodedDs(self.encoders, dev_data, self.target)
train_data = EncodedDs(self.encoders, train_data, self.target)
# --------------- #
# Update/Adjust Mixers
# --------------- #
log.info("Updating the mixers")
for mixer in self.mixers:
mixer.partial_fit(train_data, dev_data, adjust_args)
@timed_predictor
def predict(self, data: pd.DataFrame, args: Dict = {}) -> pd.DataFrame:
self.mode = "predict"
n_phases = 3 if self.pred_args.all_mixers else 4
if len(data) == 0:
raise Exception(
"Empty input, aborting prediction. Please try again with some input data."
)
self.pred_args = PredictionArguments.from_dict(args)
log.info(f"[Predict phase 1/{n_phases}] - Data preprocessing")
if self.problem_definition.ignore_features:
log.info(f"Dropping features: {self.problem_definition.ignore_features}")
data = data.drop(
columns=self.problem_definition.ignore_features, errors="ignore"
)
for col in self.input_cols:
if col not in data.columns:
data[col] = [None] * len(data)
# Pre-process the data
data = self.preprocess(data)
# Featurize the data
log.info(f"[Predict phase 2/{n_phases}] - Feature generation")
encoded_ds = self.featurize({"predict_data": data})["predict_data"]
encoded_data = encoded_ds.get_encoded_data(include_target=False)
log.info(f"[Predict phase 3/{n_phases}] - Calling ensemble")
@timed
def _timed_call(encoded_ds):
if self.pred_args.return_embedding:
embedder = Embedder(self.target, mixers=list(), data=encoded_ds)
df = embedder(encoded_ds, args=self.pred_args)
else:
df = self.ensemble(encoded_ds, args=self.pred_args)
return df
df = _timed_call(encoded_ds)
if not (
any(
[
self.pred_args.all_mixers,
self.pred_args.return_embedding,
self.problem_definition.embedding_only,
]
)
):
log.info(f"[Predict phase 4/{n_phases}] - Analyzing output")
df, global_insights = explain(
data=data,
encoded_data=encoded_data,
predictions=df,
ts_analysis=None,
problem_definition=self.problem_definition,
stat_analysis=self.statistical_analysis,
runtime_analysis=self.runtime_analyzer,
target_name=self.target,
target_dtype=self.dtype_dict[self.target],
explainer_blocks=self.analysis_blocks,
pred_args=self.pred_args,
)
self.global_insights = {**self.global_insights, **global_insights}
self.feature_cache = (
dict()
) # empty feature cache to avoid large predictor objects
return df
def test(
self,
data: pd.DataFrame,
metrics: list,
args: Dict[str, object] = {},
strict: bool = False,
) -> pd.DataFrame:
preds = self.predict(data, args)
preds = preds.rename(columns={"prediction": self.target})
filtered = []
# filter metrics if not supported
for metric in metrics:
# metric should be one of: an actual function, registered in the model class, or supported by the evaluator
if not (
callable(metric)
or metric in self.accuracy_functions
or metric in mdb_eval_accuracy_metrics
):
if strict:
raise Exception(f"Invalid metric: {metric}")
else:
log.warning(f"Invalid metric: {metric}. Skipping...")
else:
filtered.append(metric)
metrics = filtered
try:
labels = self.model_analysis.histograms[self.target]["x"]
except:
if strict:
raise Exception("Label histogram not found")
else:
label_map = (
None # some accuracy functions will crash without this, be mindful
)
scores = evaluate_accuracies(
data,
preds[self.target],
self.target,
metrics,
ts_analysis=self.ts_analysis,
labels=labels,
)
# TODO: remove once mdb_eval returns an actual list
scores = {k: [v] for k, v in scores.items() if not isinstance(v, list)}
return pd.DataFrame.from_records(
scores
) # TODO: add logic to disaggregate per-mixer
As you can see, an end-to-end pipeline of our entire ML procedure has been generating. There are several abstracted functions to enable transparency as to what processes your data goes through in order to build these models.
The key steps of the pipeline are as follows:
Run a statistical analysis with
analyze_dataClean your data with
preprocessMake a training/dev/testing split with
splitPrepare your feature-engineering pipelines with
prepareCreate your features with
featurizeFit your predictor models with
fit
You can customize this further if necessary, but you have all the steps necessary to train a model!
We recommend familiarizing with these steps by calling the above commands, ideally in order. Some commands (namely prepare, featurize, and fit) do depend on other steps.
If you want to omit the individual steps, we recommend your simply call the learn method, which compiles all the necessary steps implemented to give your fully trained predictive models starting with unprocessed data!
6) Call python to run your code and see your preprocessed outputs
Once we have code, we can turn this into a python object by calling predictor_from_code. This instantiates the PredictorInterface object.
This predictor object can be then used to run your pipeline.
[8]:
# Turn the code above into a predictor object
predictor = predictor_from_code(code)
[9]:
# Pre-process the data
cleaned_data = predictor.preprocess(data)
train_test_data = predictor.split(cleaned_data)
INFO:dataprep_ml-3005:Cleaning the data
DEBUG:lightwood-3005: `preprocess` runtime: 18.57 seconds
INFO:dataprep_ml-3005:Splitting the data into train/test
DEBUG:lightwood-3005: `split` runtime: 1.61 seconds
[10]:
plt.rcParams['font.size']=15
f = plt.figure(figsize=(18, 5))
ax = f.add_subplot(1,3,1)
ax.hist(train_test_data["train"]['Class'], bins = [-0.1, 0.1, 0.9, 1.1], log=True)
ax.set_ylabel("Log Counts")
ax.set_xticks([0, 1])
ax.set_xticklabels(["0", "1"])
ax.set_xlabel("Class")
ax.set_title("Train:\nDistribution of Classes")
ax.set_ylim([1, 1e6])
ax = f.add_subplot(1,3,2)
ax.hist(train_test_data["dev"]['Class'], bins = [-0.1, 0.1, 0.9, 1.1], log=True, color='k')
ax.set_ylabel("Log Counts")
ax.set_xticks([0, 1])
ax.set_xticklabels(["0", "1"])
ax.set_xlabel("Class")
ax.set_title("Dev:\nDistribution of Classes")
ax.set_ylim([1, 1e6])
ax = f.add_subplot(1,3,3)
ax.hist(train_test_data["test"]['Class'], bins = [-0.1, 0.1, 0.9, 1.1], log=True, color='r')
ax.set_ylabel("Log Counts")
ax.set_xticks([0, 1])
ax.set_xticklabels(["0", "1"])
ax.set_xlabel("Class")
ax.set_title("Test:\nDistribution of Classes")
ax.set_ylim([1, 1e6])
f.tight_layout()
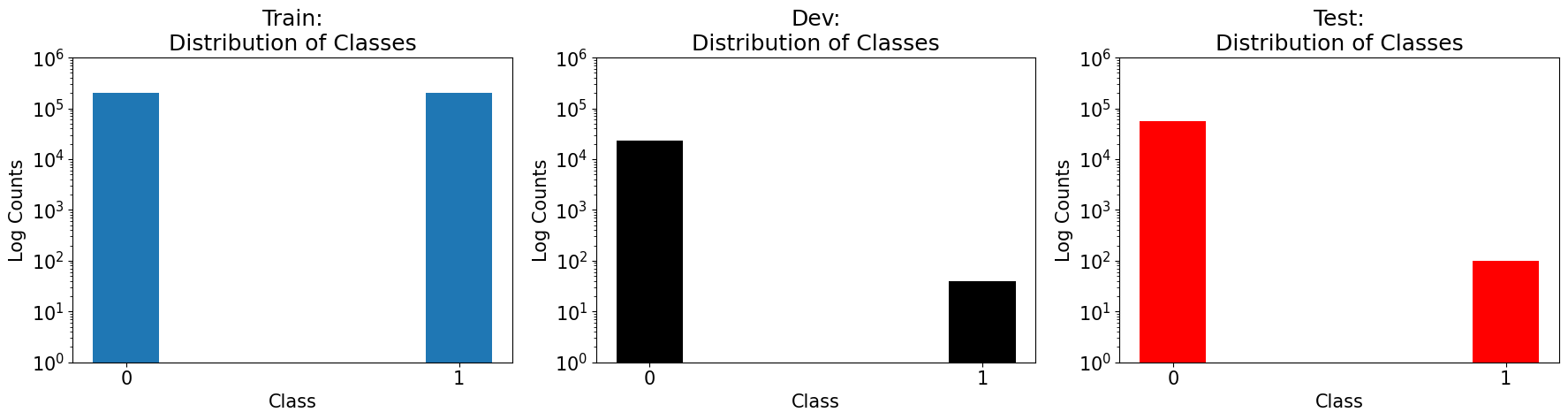
As you can see, our splitter has greatly increased the representation of the minority class within the training data, but not so for the testing or dev data.
We hope this tutorial was informative on how to introduce a custom splitter method to your datasets! For more customization tutorials, please check our documentation.
If you want to download the Jupyter-notebook version of this tutorial, check out the source github location found here: lightwood/docssrc/source/tutorials/custom_splitter.